Introduction
Total Internal Reflection Fluorescence Microscopy is a single molecule imaging technique which allows for precise observations of single molecule binding events to a surface. TIRF microscropy can be used to provide data on how our HIV-1 CA proteins bind to our DNA origami scaffold structure.
Our microscope is able to execute fluorescent microscopy in two settings, epifluorescence microscopy and TIRF microscopy. The difference between these settings is the angle at which the laser is directed towards the objective (see figure 1). Epifluorescence allows a directed column of laser to pass through the entirety of the sample, whereas TIRF angles the laser to a reflective angle, with the site of the angle of incidence emitting an evanescent field which penetrates only a thin layer of the sample due to degradation of the light over time.
TIRF microscopy is ideally suited to single molecule experiments as the illumination of only a thin layer of sample allows for the elimination of noise caused by excess fluorophores, as only fluorophores bound to the surface will emit an excitation signal.
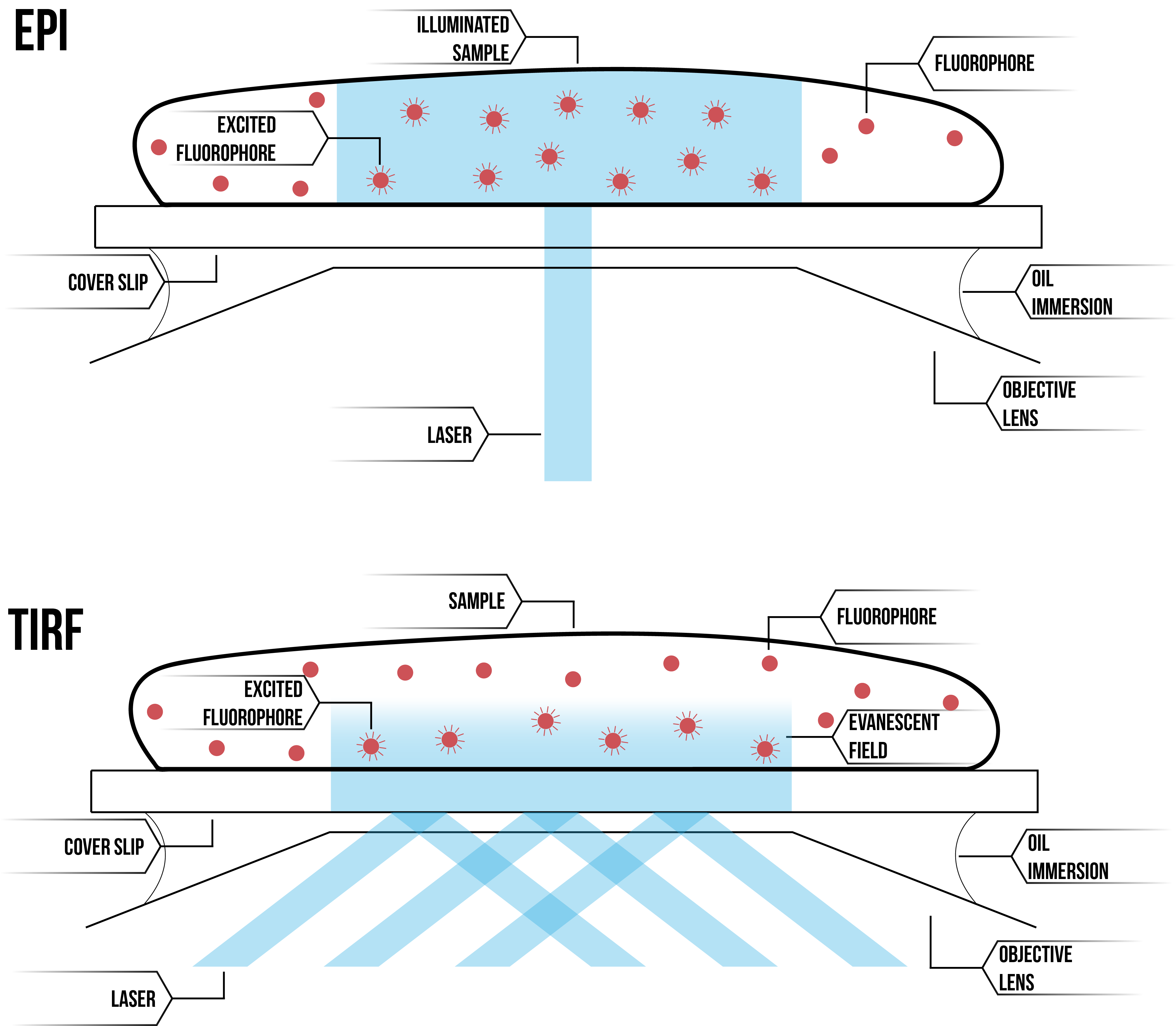
Figure 1: Comparative diagram of epifluorescence microscopy (top) and TIRF microscopy (bottom).
As TIRF Microscopy allows the observation of single molecule binding events, it would be an ideal technique for both qualitatively and quantitatively measuring protein binding events. However, due to the short time frame of the BIOMOD competition, we were unable to reach this point in analysis. This would be a beneficial future direction for the project, with our preliminary experiments having set the ground-work for such experiments.
Aim
TIRF experiments aim to confirm the functionality of our DNA origami structure as a scaffold for protein localisation.
The Microscope
The Lee Lab in which we have been completing our project has recently set up a new TIRF microscope for use in single molecule experiments. We have spent time aiding in microscope set-up, familiarising ourselves with the machinery, and testing the imaging programs in preparation for future use. See below Figures 2 and 3 detailing the set-up of our microscope.
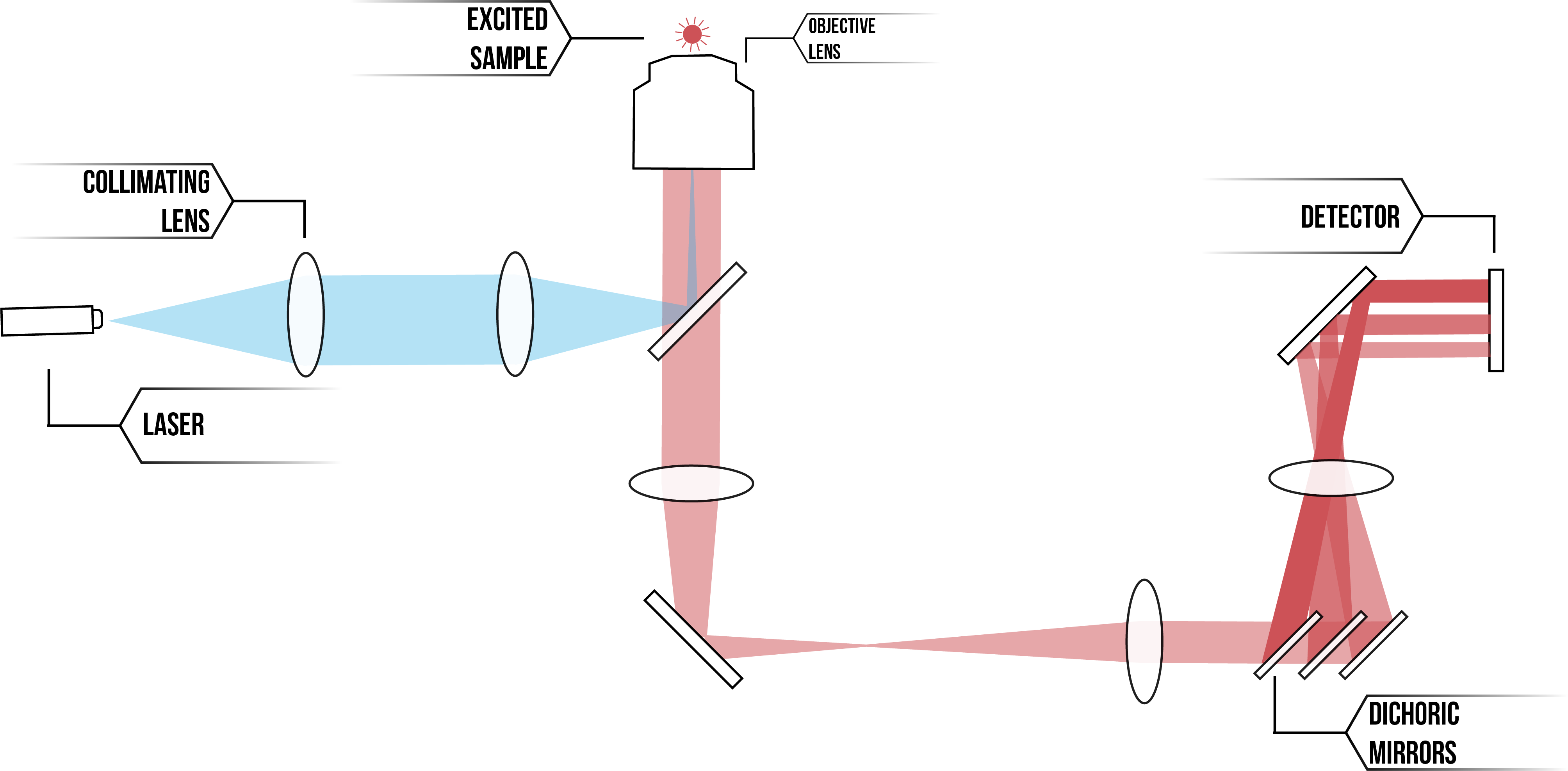
Figure 2: Schematic of TIRF microscope set-up

Figure 3: Images of Microscope
Production of Materials
In order to observe individual binding events of proteins to our DNA origami structure, an initial layer of surface chemistry must be immobilised as a binding surface. We have identified and practiced preparation of a BSA/biotin/streptavidin conjugation system for use in our future experiments. We have also produced our DNA origami structure with biotinylated extensions, for the purpose of fixing to our cover slip surface, and conjugated HIV-1 CA proteins with fluorophores in preparation for imaging. Unfortunately, due to time constraints and the technical inefficiencies posed by using a new microscope for the first time, we were unable to conduct our proposed single molecule experiments.
Future Directions
Characterisation of protein binding sites using PAINT
Point Accumulation in nanoscale topography (PAINT) is a method which uses transient binding events of short fluorophore labelled oligonucleotides, imager strands, to single stranded extensions of an origami structure, docking strands, to generate a blinking signal. The blinking effect produced by this technique is used for super-resolution reconstruction of the location of binding events, therefore indicating the precise location of the binding sites on our DNA origami structure by stochastic imaging.
We propose to use PAINT as a method to characterise our DNA origami scaffold. The process of imager strands binding and unbinding from the extension strands of our structure will enable us to assess the functionality of our scaffold. We will generate information about the functionality of these sites to bind ssDNA-conjugated proteins, as well as information about the distances between binding sites, indicating the ability of the structure to influence protein-protein interactions.
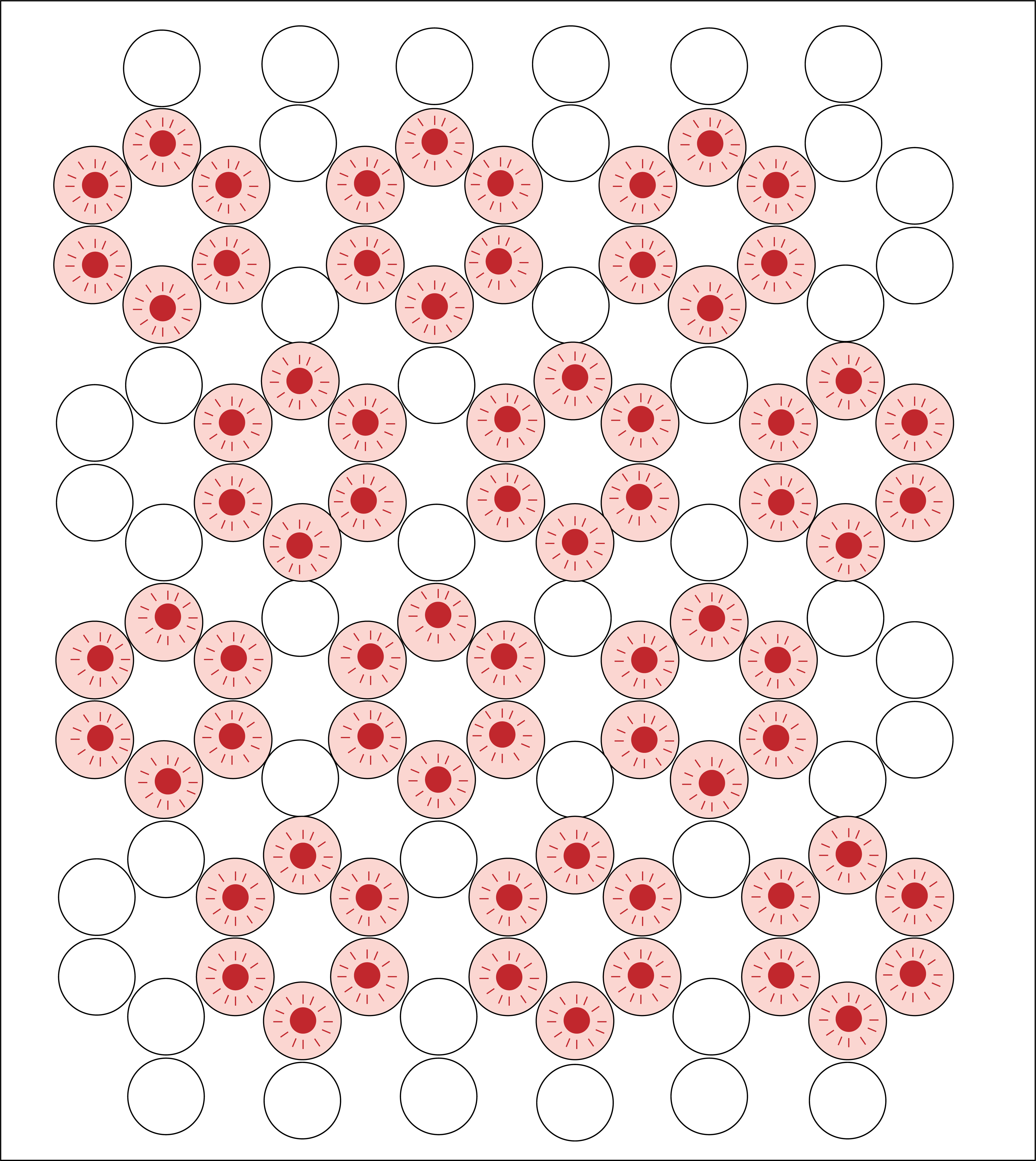
Figure 4: Diagram of proposed PAINT experiment to characterise our DNA origami structure. Binding sites of DNA origami indicated in pink, excited fluorophores indicated in red.
Seeded assembly of HIV-1 CA proteins
Extending upon characterisation of our DNA origami scaffold, we would like to show how HIV-1 CA proteins are able to bind to the scaffold, and by localisation begin to bind to each other and assemble into hexameric lattice structures. This would be accurately achievable using TIRF microscopy, as elimination of background noise will enable extraction of accurate kinetic data.
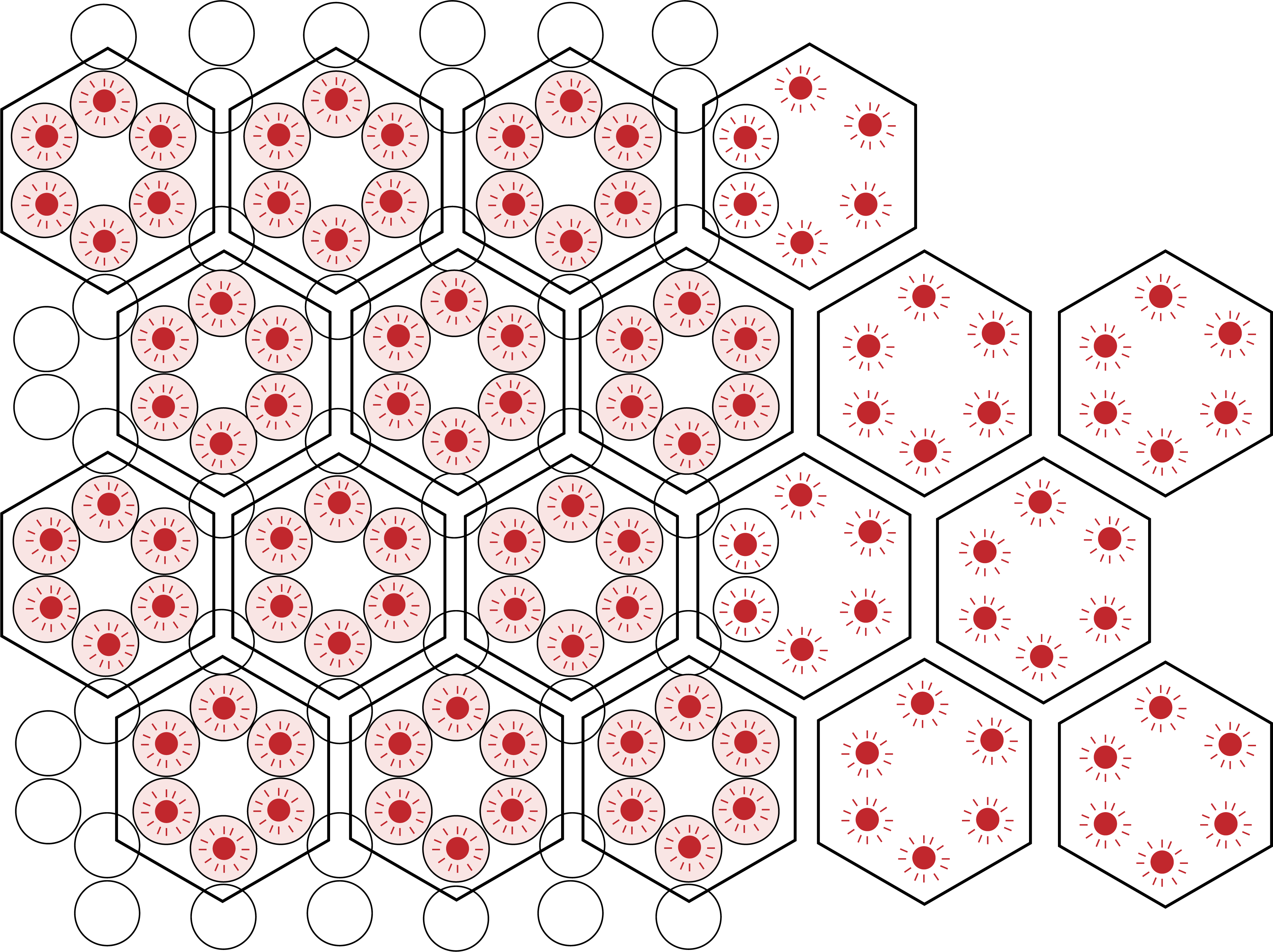
Figure 5: Diagram of proposed seeding experiment to characterise latticing of HIV-1 CA proteins. DNA origami represented as honeycomb lattice, with binding sites of DNA origami indicated in pink. Excited, protein bound fluorophores indicated in green, and extension of such hexameric proteins beyond the scaffold showing how assembly may occur.
References
- Fish KN. Total Internal Reflection Fluorescence (TIRF) Microscopy. Current protocols in cytometry / editorial board, J Paul Robinson, managing editor . [et al]. 2009;0 12:Unit12.18.
- Schnitzbauer J, Strauss MT, Schlichthaerle T, Schueder F, Jungmann R. Super-resolution microscopy with DNA-PAINT. Nat Protoc. 2017;12(6):1198-1228.
- Chen B, Tycko R. Simulated Self-Assembly of the HIV-1 Capsid: Protein Shape and Native Contacts Are Sufficient for Two-Dimensional Lattice Formation. Biophysical Journal. 2011;100(12):3035-3044.